News
2026
- A new undergraduate student Arjun Rajagopal joins the lab. Welcome Arjun!
2025
- Marcel Goldschen-Ohm was elected to the Society of General Physiologists council.
- Marcel Goldschen-Ohm and Blair Johnson were awarded an APX grant from the University of Texas at Austin to develop an automated patch clamp combining custom fluidics with common electrophysiology equipment to enable rapid ion channel variant diagnosis and drug screening at low cost for individual research labs.
- Marcel Goldschen-Ohm and Andres Jara-Oseguera were awarded a Catalyst 2.0 grant from the University of Texas at Austin College of Natural Sciences to lay the foundation for a new collaborative project designed to screen every possible single residue variant in a synpatic GABAA receptor with a high throughput unbiased approach involving fluorescence detection of channel activity and next generation sequencing.
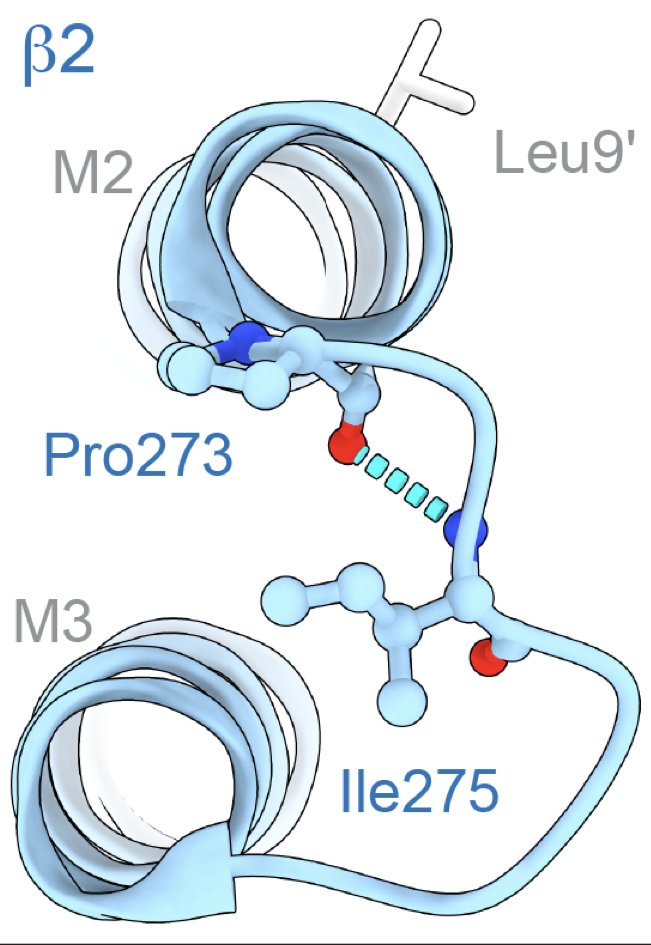
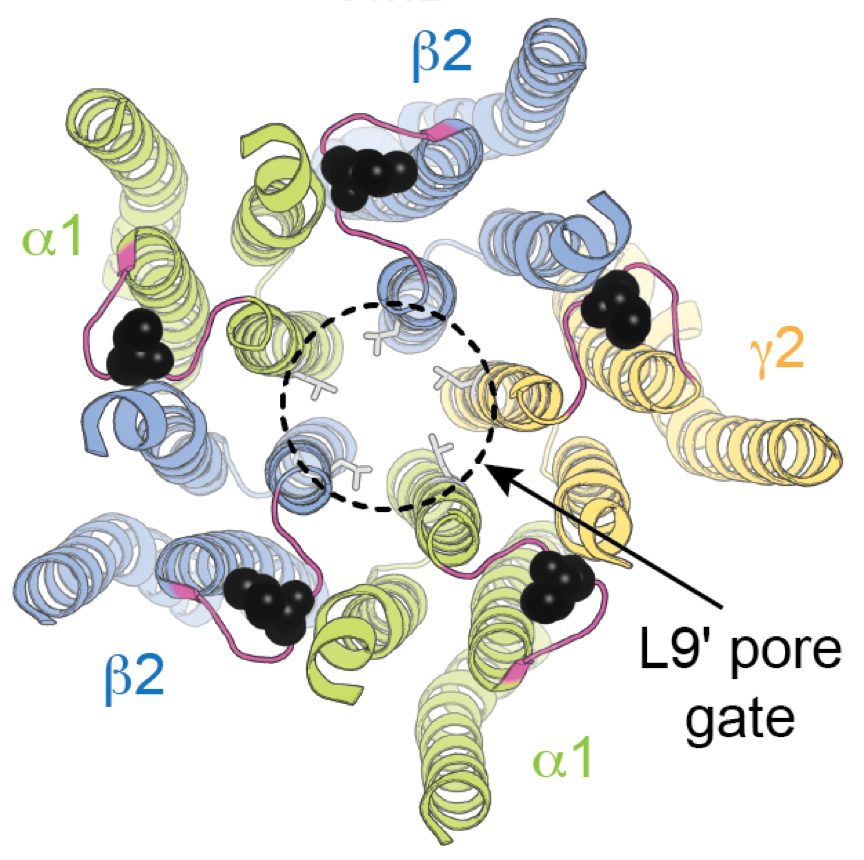
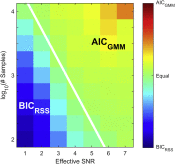
- Cecilia Borghese and coworkers uncover a new part of the activation mechanism coupling binding to gating in GABAA receptors that involves backbone hydrogen bonding resistant to genetic mutation. Read about it in Nature Communications. This work was a collaboration with Jason Galpin and Chris Ahern from the University of Iowa and Samuel Eriksson Lidbrink, Reba Howard, and Erik Lindahl from Stockholm University.
- Darren Lopez recieves an Outstanding Teaching Assistant Award for his phenomenal job as an undergraduate teaching assistant in Marcel Goldschen-Ohm's course Programming and Data Analysis for Modern Neuroscience.
- Khadeeja Shah wins an award for her outstanding poster presentation of her research at the Technology & Science Undergraduate Research Forum.
- Netrang Desai was awarded an Undergraduate Summer Training Fellowship for his project on GABAA receptor structure-function.
- Marcel Goldschen-Ohm has been promoted to Associate Professor with Tenure.
- Marcel Goldschen-Ohm presents the lab's latest work in a special symposium "Best of Biophysics" at the Biophysical Society annual meeting.
- Our recent paper was selected as the Paper of the Year by the Biophysical Journal and Biophysical Society which annually recognizes a single outstanding paper from an early career investigator. For this award, Marcel Goldschen-Ohm was invited to speak in the Best of Biophysical Journal symposium at the Biophysical Society annual meeting.
- A new undergraduate student Zaid Ahmed joins the lab. Welcome Zaid!
2024
- Cecilia Borghese and coworkers uncover a new part of the activation mechanism coupling binding to gating in GABAA receptors that involves backbone hydrogen bonding resistant to genetic mutation. Read about it in bioRxiv.
- Marcel hosts Jorge Contreras for the department of Neuroscience seminar series at the University of Texas at Austin.
- Marcel hosts Chen Zhao for the department of Neuroscience seminar series at the University of Texas at Austin.
- A new undergraduate student Khadeeja Shah joins the lab. Welcome Khadeeja!
- Netrang Desai was awarded an Undergraduate Research Fellowship for his project on GABAA receptor structure-function.
- Marcel Goldschen-Ohm was selected to receive a 2024 College of Natural Sciences Teaching Excellence Award for his outstanding teaching in the Department of Neuroscience at the University of Texas at Austin.
- Marcel Goldschen-Ohm presents Cecilia Borghese's work on backbone hydrogen bonding in GABAA receptor activation at the Ion Channels Gordon Research Conference. This work was a collaboration with Jason Galpin and Chris Ahern from the University of Iowa and Samuel Lidbrink, Reba Howard, and Erik Lindahl from Stockholm University.
- Marcel Goldschen-Ohm and Baron Chanda wrote an editorial for the special issue Bioelectricity and Molecular Signaling in honor of Richard Aldrich. Read about it in Biophysical Journal
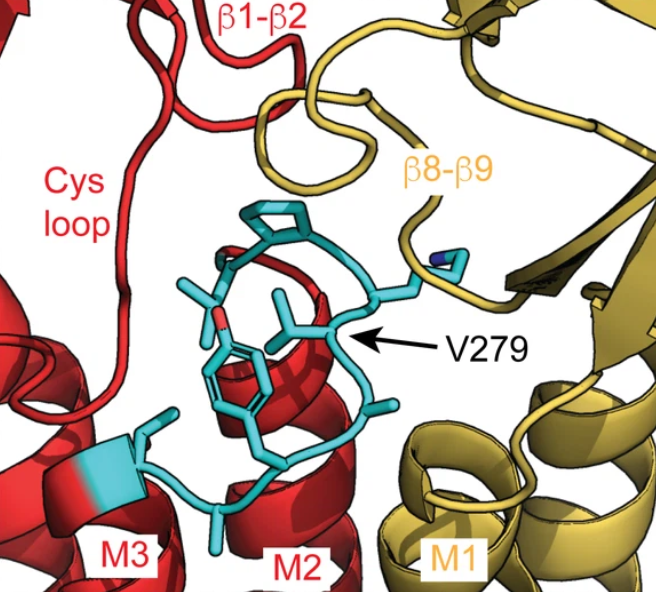
- Zachary Endres and Joseph (Wagner) Nors show that the central hydrophobic residues in GABAA receptor M2-M3 linkers have highly asymmetric subunit-specific roles in gating and drug modulation. Read about it in Biophysical Journal
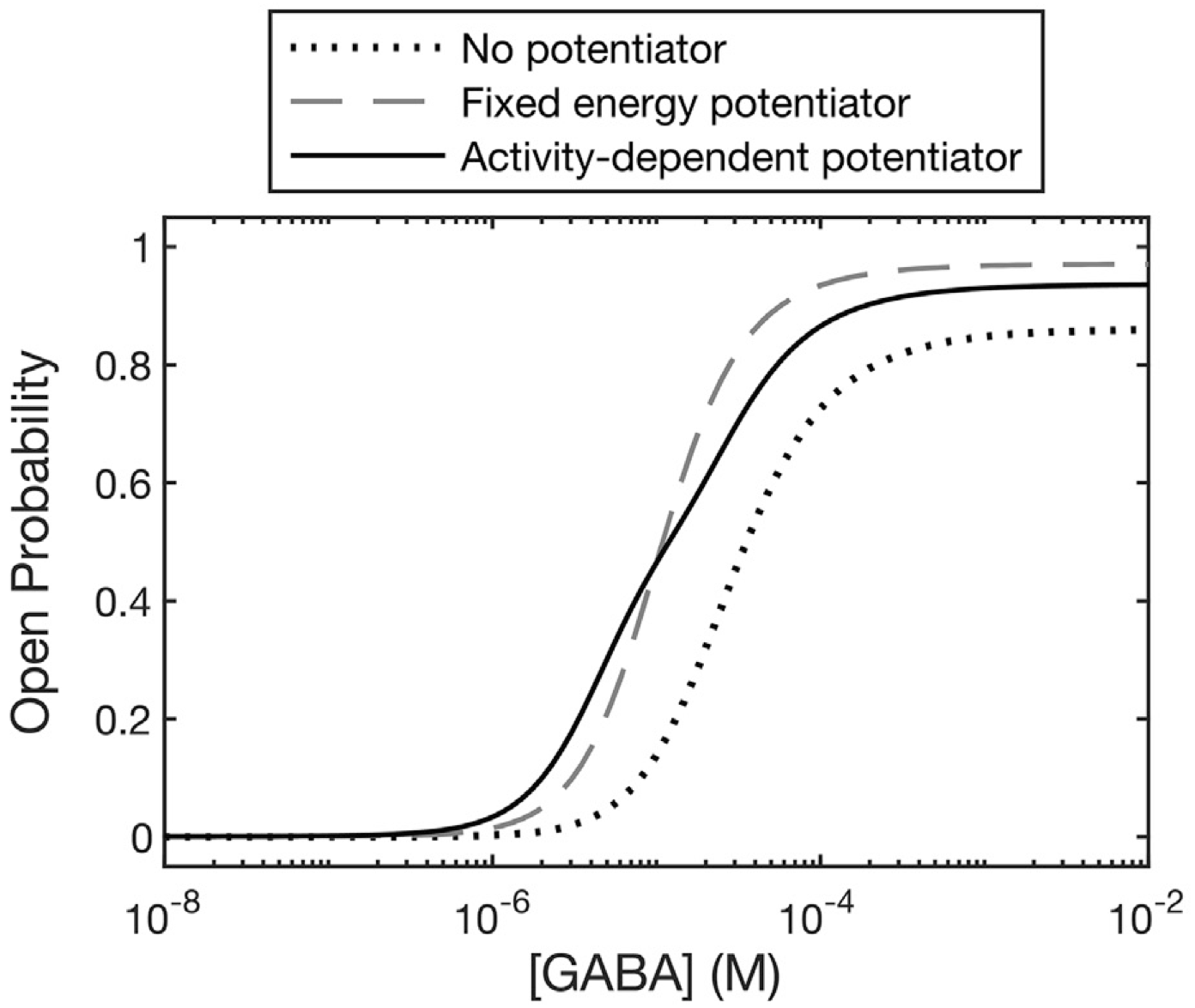
- Cecilia Borghese and Marcel Goldschen-Ohm discuss a new and notable article on the energetics of GABAA receptor allosteric potentiators. Read about it in Biophysical Journal
- Cecilia Borghese presents her work on backbone hydrogen bonding in GABAA receptor activation at the Biophysical Society 2024 annual meeting.
- New undergraduate students Rohith Gajjala, Vignesh Sadasivan, and Vineeth Murugan join the lab. Welcome Rohith, Vignesh, and Vineeth!
2023
- A new undergraduate student Netrang Desai joins the lab. Welcome Netrang!
- A new postdoc Tapas Haldar joins the lab. Welcome Tapas!
- Marcel Goldschen-Ohm was invited to speak at the Sophion Bioscience Ion Channel Modulation Symposium in Irvine, CA on October 18-19 2023.
- Marcel Goldschen-Ohm serves as editor for a special issue in Biophysical Journal honoring the scientific contributions of Richard Aldrich: Bioelectricity and Molecular Signaling.
- A new graduate student Myles Joyce rotates in the lab. Welcome Myles!
- A new research scientist Cecilia Borghese joins the lab. Welcome Cecilia!
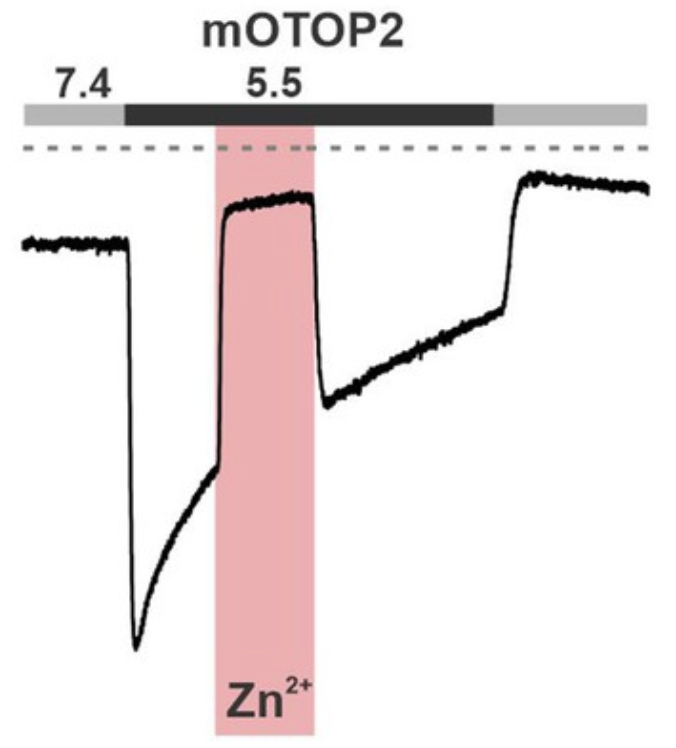
- In collaboration with Emily Liman we investigate gating of OTOP sour tast receptors by Zinc. Read about it in eLife
- Marcel Goldschen-Ohm's review of the molecular mechanisms of benzodiazepines was chosen as an Editor's Choice Article in the journal Biomolecules for its high quality and high amount of downloads.
- Marcel Goldschen-Ohm gives invited seminar at the University of Iowa Deparment of Molecular Physiology & Biophysics.
- Marcel Goldschen-Ohm invited to speak at the Biophysical Society 2023 annual meeting workshop on high-throughput single-molecule spectroscopy.
- A new postdoc Zubayer Ibne Ferdous joins the lab. Welcome Zubayer!
2022
- The lab was awarded an R01 from the NIH.
- The lab was awarded an R03 from the NIH.
- Marcel Goldschen-Ohm reviews molecular mechanisms of benzodiazepines, one of the most widely prescribed class of psycotropic drugs today. Read about it in Biomolecules.
- Marcel Goldschen-Ohm joins the editorial board at Frontiers in Pharmacology.
- Marcel Goldschen-Ohm joins the steering group at Biophysics Colab.
- Started a new collaboration with Dr. Susanne Ressl that I'm super excited about!
- Thomas Middendorf and Marcel Goldschen-Ohm discuss a new article from Godellas and Grosman about the surprising difficulty of equilibrium binding measurements for ligand-gated ion channels. Read about it in the Journal of General Physiology.
- Argha Bandyopadhyay awarded the prestigious honor of becoming a Deans Honored Graduate, a distinction only given to a few of the very top students at the University.
- Marcel Goldschen-Ohm May 5th seminar for UT Biochemistry.
- Marcel Goldschen-Ohm invited to speak at the Ion Channels 2022 Gordon Research Conference.
- Joseph (Wagner) Nors presents his work on drug modulation in GABAA receptors in a platform presentation at the Biophysical Society 2022 annual meeting. Vishal Patel and Argha Bandyopadhyay also give poster presentations of their work at the meeting.
2021
- Khue Tran and Argha Bandyopadhyay develop smBEVO which leverages computer vision algorithms for best in class baseline drift correction in single-molecule time series. Read about it on bioRxiv
- Vishal Patel, Mohamed Salinas, Darong Qi and coworkers use cell-derived nanovesicles and single-molecule optical tracking of individual ligand binding events to show that cyclic nucleotide binding in a CNG channel is followed by a conformational change of the binding domain(s) that preceeds pore opening. These observations also reveal that binding of the second of four cyclic nucleotides is either independent of the first or at most a weakly positive cooperative process. Read about it in Nature Communications and also check out the Biophysics Colab peer reviews
- Argha Bandyopadhyay spearheads development of AutoDISC which optimizes DISC for completely automated model-free idealization of single-molecule time series with state-of-the-art accuracy and performance. Read about it in Biophysical Journal and download AutoDISC for MATLAB
- Khue Tran recieves TIDES fellowship for summer research.
- Joseph (Wagner) Nors and coworkers identify a critical residue mediating coupling between the benzodiazepine drug site and the channel pore in a neurotransmitter-gated GABAA receptor. Read about it in eLife
- Argha Bandyopadhyay presents a video and poster at the Capital of Texas Undergraduate Research Conference about his work on a novel algorithm for automated single-molecule time series analysis. Watch the video
2020
- Marcel Goldschen-Ohm joins the board of reviewing editors at eLife.
- Joseph (Wagner) Nors presents a poster at the Biophysical Society annual meeting about his work on coupling between the benzodiazepine binding site and channel pore in GABAA receptors.
- Argha Bandyopadhyay presents a poster at the American Physician Scientists Association (APSA) conference about his work on a novel algorithm for automated single-molecule time series analysis.
- David White spearheads developement of DISC for model-free idealization of single-molecule time series with state-of-the-art accuracy and performance. Read about it in eLife
2019
- Rocio Finol-Urdaneta and coworkers show that batrachotoxin props open the pore of prokaryotic voltage-gated sodium channels. Read about it in the Journal of General Physiology
Research
Ion channels are critical for rapid signaling between neurons and other excitable cells throughout the body. Drug binding to specific recognition sites allows tuning channel behavior to treat aberrant behavior associated with human disorders. An overarching goal of the lab is to understand the molecular mechanisms by which channels operate and the physical basis by which small molecule ligand/drug binding regulates their activity. Understanding these mechanisms will provide a necessary framework for rational development of novel therapeutic strategies to improve treatments that impact human health.
The lab utilizes a variety of approaches such as fluorescence spectroscopy, electrophysiology and biochemistry at both single-molecule and ensemble levels to probe the dynamic motions of proteins that underlie their biological function. As part of this process, we are also interested in developing new approaches or workflows to resolve the molecular forces and interactions involved in drug binding and modulation.
Ion channels are natures transistors/transducers.
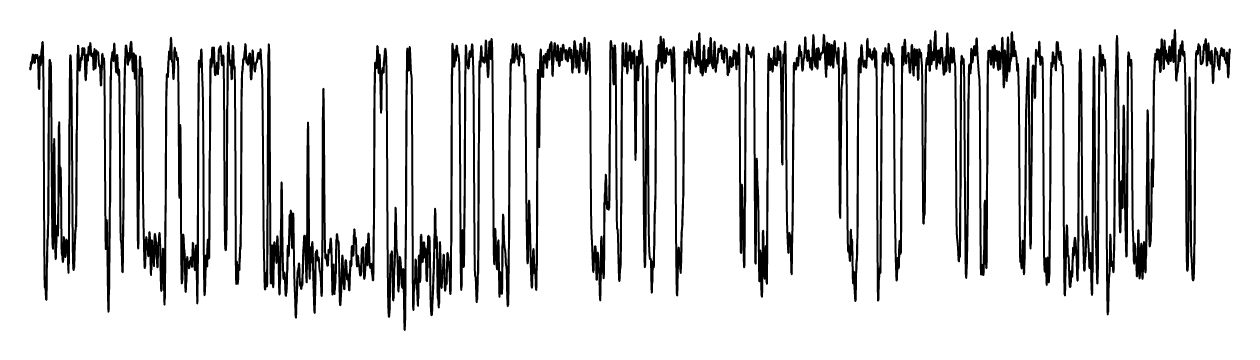
Single-molecule fluorescence imaging.

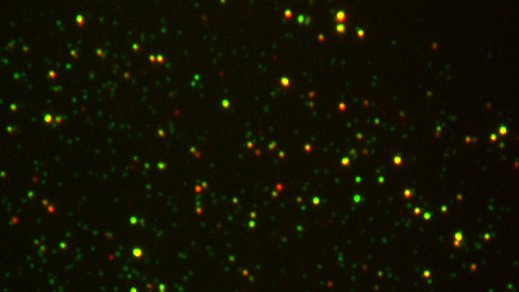
- Cell-derived nanovesicles for single-molecule fluorescence
- Single-molecule ligand binding dynamics
- FRET-based approaches to probe conformational dynamics
- Unnatrual Amino Acids
- Click Chemistry for site-specific fluorescence
- Cryo-EM structural analysis of ion channels
People






Past Lab Members
Joined the Sun lab at the University of Texas Medical Branch

Joined the Fox and Baratono lab in the Center for Brain Circuit Therapeutics at Brigham and Women's Harvard Medical School

Joined the Miller School of Medicine at the University of Miami

UT Austin Deans Honored Graduate
Applying to medical school

Joined the Goodman lab in the Molecular and Cellular Physiology PhD program at Stanford

UT Austin Deans Honored Graduate
Joined the Williams lab in the MD/PhD program at NYU Langone

Joined Lawrence Berkeley National Laboratory

Joined the MD program at Dell Medical School at the University of Texas at Austin
Residency at Mass General in Boston

Joined the Neurobiology and Behavior PhD program at Stony Brook University
Software
- napari-cosmos-ts: Napari plugin for colocalization single-molecule spectroscopy (CoSMoS) time series (TS) analysis. Find molecules and extract and visualize their one-dimensional time series.
- xarray-graph: GUI for visualizing and analyzing one-dimensional time series slices from a tree of xarray datasets. Includes regions, measurement, curve fitting, and more.
- pyDISC: GUI for idealizing one-dimensional time series using the DIvisive Segmentation and Clustering (DISC) algorithm plus tools for adjusting number of levels or generic HMM optimizaiton. An excellent tool for idealizing time series, tested primarily on fluorescence data. In many cases automated idealization is very good, but otherwise manual refinement by adding/removing levels and/or HMM optimization does the trick.
- AutoDISC: MATLAB UI extending the DIvisive Segmentation and Clustering (DISC) algorithm for completely automated model-free idealization of single-molecule piecewise constant time series in the presence of per-molecule variability as often observed in high-throughput fluorescence imaging paradigms such as TIRF. Read about it in Biophysical Journal
- smBEVO: Single-molecule time series baseline estimation via visual optimization. smBEVO leverages computer vision algorithms for best in class baseline drift correction in single-molecule time series. Read about it on bioRxiv
- Kinetic Model Builder: HMM model building and optimization GUI for patch-clamp electrophysiology (or other time series) data. MacOSX only.
- EigenLab: C++ utility for parsing/evaluating matrix math equations input at run time in a format similar to MATLAB (requires Eigen for matrix math).
- single-molecule-imaging-toolbox: A suite of programs for single-molecule image analysis in MATLAB.
- PlotXRegionsUITool: MATLAB tool for interactive plot x-axis region selection and analysis/manipulation of plotted data based on selected regions. Supports plotBig_Matlab for fast plotting of large data traces (e.g. single channel records).
- PatchMeister: MATLAB graphical user interface for electrophysiology (or other time series) analysis. Note, in the future PlotXRegionsUITool will either supercede or merge with this package.
Resources
- Biophysics Colab
- Biophysical Society
- Society of General Physiologists (SGP)
- Texas Advanced Computing Center (TACC)
- GROMACS Molecular Dynamics
- Ai2 Paper Finder: Find relevant literature.
- Semantic Scholar: Find relevant literature.
- Research Rabbit: Literature mapping from seed paper.
- Elicit: Find relevant literature.
- Ai2 Scholar QA
- STORM
- SciSpace
- NotebookLM: Chat about a bunch of selected PDFs.
- Inciteful: Select two papers and we will show you how the literature connects them together.
- Paperpal: Word plugin.
- Consensus
- Julius: Analyze data.
- ThesisAI: Draft scientific documents.
- Jenny AI: Academic writing assistant.
Publications
2025
- A single main-chain hydrogen bond required to keep GABAA receptors closed. Nature Communications
2024
- Bioelectricity and molecular signaling. Biophysical Journal
- GABAA receptor subunit M2-M3 linkers have asymmetric roles in pore gating and diazepam modulation. Biophysical Journal (voted Paper of the Year by the Biophysical Society)
- State-dependent energetics of GABAA receptor modulators. Biophysical Journal
2023
- Zn2+ potentiation of OTOP proton channels identifies structural elements of the gating apparatus. eLife
2022
- Benzodiazepine modulation of GABAA Receptors: A mechanistic perspective. Biomolecules
- The surprising difficulty of "simple" equilibrium binding measurements on ligand-gated ion channels. Journal of General Physiology
2021
- smBEVO: A computer vision approach to rapid baseline correction of single-molecule time series. bioRxiv
- Single-molecule imaging with cell-derived nanovesicles reveals early binding dynamics at a cyclic nucleotide-gated ion channel. Nature Communications
- Unsupervised selection of optimal single-molecule time series idealization criterion. Biophysical Journal
- A critical residue in the alpha1 M2-M3 linker regulating mammalian GABAA receptor pore gating by diazepam. eLife
2020
2019
- Batrachotoxin acts as a stent to hold open homotetrameric prokaryotic voltage-gated sodium channels. Journal of General Physiology
Prior Publications
2017
- SnapShot: Channel Gating Mechanisms. Cell
- Observing single-molecule dynamics at millimolar concentrations. Angewandte Chemie
- The intrinsically-liganded cyclic nucleotide-binding homology domain promotes KCNH channel activation. Journal of General Physiology
2016
- Structure and dynamics underlying elementary ligand binding events in human pacemaking channels. eLife
2015
- Exocytotic fusion pores are composed of both lipids and proteins. Nature Structural & Molecular Biology
- How to open a proton pore-more than S4? Nature Structural & Molecular Biology
2014
- A nonequilibrium binary elements-based kinetic model for benzodiazepine regulation of GABAA receptors. Journal of General Physiology
- Evolutionarily conserved intracellular gate of voltage-dependent sodium channels. Nature Communications
- Probing gating mechanisms of sodium channels using pore blockers. Handbook of Experimental Pharmacology
2013
- Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. Journal of General Physiology
- Multiple pore conformations driven by asynchronous movements of voltage sensors in a eukaryotic sodium channel. Nature Communications
2011
- Three arginines in the GABAA receptor binding pocket have distinct roles in the formation and stability of agonist- versus antagonist-bound complexes. Molecular Pharmacology